Recently, scientists of YSFRI generated a chromosome-level genome assembly of the greenfin horse-faced filefish (Thamnaconus septentrionalis). The research results were published online in Molecular Ecology Resources on May 10th.
T. septentrionalis is a valuable commercial fish species that is widely distributed in the Indo-West Pacific Ocean. This fish has characteristic blue-green fins, rough skin and a spine-like first dorsal fin. An interesting feature of T. septentrionalis is its rough skin, which is observed because the skin is covered with dense small scales. These scales are difficult to remove, and people have to peel off the skin before eating. Given this property, filefish is also called “skinned fish” in China. T. septentrionalis represents a conservation issue because its population has declined sharply, and it is an important marine aquaculture fish species in China. The genomic resources of the filefish are lacking, and no reference genome has been released.
The first chromosome-level genome of T. septentrionalis was constructed using nanopore sequencing and Hi-C technology. A total of 50.95 Gb polished nanopore sequences were generated and were assembled to 474.31 Mb genome, accounting for 96.45% of the estimated genome size of this filefish. The assembled genome contained only 242 contigs, and the achieved contig N50 was 22.46 Mb, reaching a surprisingly high level among all the sequenced fish species. Hi-C scaffolding of the genome resulted in 20 pseudochromosomes containing 99.44% of the total assembled sequences. The genome contained 67.35 Mb of repeat sequences, accounting for 14.2% of the assembly. A total of 22,067 protein-coding genes were predicted, 94.82% of which were successfully annotated with putative functions. Furthermore, a phylogenetic tree was constructed using 1,872 single-copy orthologous genes and 67 unique gene families were identified in T. septentrionalis genome.
In China, the demand sizes of filefish vary in different regions, and the smallest size is only 150 g, which can be achieved within one year. One bottleneck of T. septentrionalis cultivation is the high mortality under high temperature, especially when the water temperature is above 25℃. The genomic information will enable GWAS to identify SNPs associated with fast-growth and heat-tolerance traits, benefit genome-assisted breeding programs and make a sustainable and profitable filefish cultivation industry. These data are also useful for diverse conservation applications including identifying conservation units, assessing gene flow, and detecting local adaptation of the populations along the coast of China and Korea. Moreover, the genome assembly and applied methodology supply valuable resources for comparative genomics studies, especially for the vertebrate and fish genomics communities.
YSFRI scientists Bian Li, Li Fenghui and Ge Jianlong are the co-first authors. YSFRI experts Chen Siqng, Shao Changwei and Lin Zhishu from Qingdao Municipal Ocean Technology Achievement Promotion Center are the co-corresponding authors.
Full text link:https://onlinelibrary.wiley.com/doi/10.1111/1755-0998.13183.

FIGURE 1 The greenfin horse-faced filefish (Thamnaconus septentrionalis)
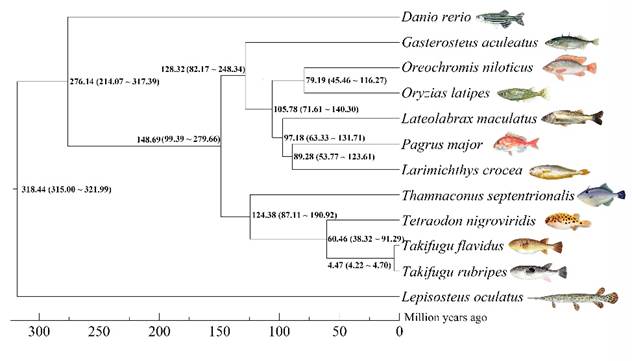
FIGURE 2 Phylogenetic analysis of the filefish with other teleost species. Lepisosteus oculatus was used as the outgroup.
